This document explains how to use several existing Soaplab clients, and/or it gives pointers where to find more about them. Before diving into details it is recommended to read a general introduction to Soaplab clients (a page you have probably just came from).
Soaplab has several simple clients that can be downloaded and immediately used. These clients are available as command-line tools, there is no GUI - their primary purpose is to test deployed services.
- Downolad soaplab-clients-YYYY-MM-DD.tar.gz from the
distribution site directory.
- Unpack it. It creates directory soaplab-clients-YYYY-MM-DD.
Go into this directory.
- There is a README file with
some instructions. The provided scripts in the run directory
take care about setting CLASSPATH and starting a client. More
about individual clients below.
These clients can be started by invoking scripts from the run directory. For example:
run/run-listThe same clients are also accessible when you use Soaplab from a CVS, or when you have installed a Soaplab binary distribution. The only difference is that, more or less from the historical reasons, they may have sometimes different names and locations (but their functionality is exactly the same because they use the same Java classes at the end).
The examples and documentation below are using names from the Soaplab client distribution. If you call clients using a different distribution note that:
- for Soaplab binary distribution the clients are in the
ws sub-directory,
- for Soaplab CVS the clients are in the run
sub-directory.
A Java client accessing an Analysis Factory Web Service that produces a list of available services. Type:
run/run-list -helpThe all options are described in a help file. The most important ones are those specifying where is the service located (hostname, port number, or the full endpoint). To access a list service running at EBI you may try to use:
run/run-list -e http://www.ebi.ac.uk/soaplab/services -a run/run-list -e http://www.ebi.ac.uk/soaplab/services -a -gowlabYou will get names of all available services. These are the names you need to know when invoking the Analysis client.
The most powerful (meaning: having the most options) Java client is an AnalysisClient that accesses individual Analysis Web Services. Type:
run/run-analysis -helpThe all options (and there are many of them) are listed in a help file. To access services running at EBI you may try to use an endpoint http://www.ebi.ac.uk/soaplab/services.
Here are some real examples to try:
run/run-analysis \
(1) -e http://www.ebi.ac.uk/soaplab/services \
(2) -name edit.seqret \
(3) sequence_direct_data aaaaaccccc \
(4) osformat embl \
(5) -w \
(6) -r
You will get hopefully something like this.
run/run-analysis \
(1) -e http://www.ebi.ac.uk/soaplab/services \
(2) -name edit.seqret \
(3) -i \
(4) -o \
(5) -t \
run/run-analysis \
(1) -e http://www.ebi.ac.uk/soaplab/services \
(2) -name edit.seqret \
(3) -ii \
(4) -oo
There are also few clients that have little real usage because they have hard-coded input data that cannot be changed (without re-compiling Java classes). They can be used, however, as quick checking whether a particular service location (an endpoint) is accessible and live. Also, they are used (in the next chapter) as code examples. Type:
run/run-test-seqret -e http://www.ebi.ac.uk/soaplab/services run/run-test-cpgplot -e http://www.ebi.ac.uk/soaplab/servicesOr, use a script to test (almost) all EMBOSS applications introduced in the EMBOSS Tutorial:
run/run-tutorialIf successful you should get something like this:
Accessing services at: http://www.ebi.ac.uk/soaplab/services/ (detail reports are in /tmp/EmbossTutorialTest.report) display.showdb.derived OK edit.seqret.derived OK OK Created: /tmp/ops2.fasta information.infoseq.derived OK OK alignment_dot_plots.dottup.derived OK alignment_global.needle.derived OK alignment_local.water.derived OK nucleic_gene_finding.getorf.derived OK nucleic_gene_finding.plotorf.derived OK nucleic_translation.transeq.derived OK Created: /tmp/xlrhodop.pep protein_composition.pepinfo.derived OK protein_2d_structure.tmap.derived OK protein_motifs.patmatmotifs.derived OK protein_motifs.pscan.derived OK alignment_multiple.emma.derived OK Created: /tmp/ops2.aln alignment_multiple.prettyplot.derived OK nucleic_profiles.prophecy.derived OK Created: /tmp/ops2.prophecy nucleic_profiles.prophet.derived OK information.tfm.derived OK
There are also two advanced clients, meant to be primarily used by the Soaplab service providers. The first one helps with the deployment of services in the Tomcat/Axis environment, the second supports automatic registration of the deployed services by some third-party service registry.
There is a special guide for Soaplab service providers. Read it first if the following text does not make much sense to you.
One of the tasks of service providers is to deploy Analysis Web Services in a Tomcat/Axis server. The deployment consists of:
That all can be done by an administrative client AAdmin.
This client should be used only on the machine where the Analysis Web Services are being deployed (because some functions would not work when invoked remotely, from the security point of view).
Just type:
run/run-admin -helpand you will get a long help description.
Note that not all functions can be invoked within the same invocation (for example, to get WSDL from running Axis, you need first to restart the servlet engine (Tomcat) after creating and compiling Java classes for derived services). Here is an example of a usual scenario (which uses a lot of default values):
Where (1) deploys an Analysis Factory Service, (2) deploys all Analysis Services, (3) creates and deploys all derived services, and (4) generates WSDL files both for normal and derived services into directory metadata (which must already exist). Note that between individual steps you need to restart your servlet engine (e.g. Tomcat).(1) run/run-admin -deploylist (2) run/run-admin -deploy (3) run/run-admin -deployx (4) run/run-admin -wsdl metadata -wsdlx metadata
You may have noticed that the examples above do not show any service names. They can be used, but if they are not - as above - the Analysis Factory Service (in the example above it was deployed in the first step) is contacted and everything is done for services known to this Factory service.
All Soaplab-based services can be also interrogated about their types, descriptions and other information useful in a registration process (such as a UDDI registry).
The Soaplab provides a special-purpose client to help with the registration process. It does not register services itself but it generates either XML files or Java source code that can be used for registration. It does it by contacting a running Soaplab server, asking for description of its services and converting this information in one of several possible formats:
- XML file (an example for two services)
- Java source code to be used with UDDI4J API (an example for the same two services)
- TBD: Java/Perl source code for BioMoby
That all can be done by client RegistryAdmin. Type:
run/run-registry -helpand you will get a long help description.
Here is an example for generating an XML file for all services provided by a given provider (put all on one line):
Where (2) tells what services to include in the resulting file (in this case, all services from the given endpoint will be included), (3) indicates what format is desired, and (4) specifies a file for the result.(1) run/run-registry \ (2) -e http://www.ebi.ac.uk/soaplab/services \ (3) -format xml \ (4) -o publish_services.xml
You may have noticed that the example above does not show any service names. An another example below shows how to generate Java source code only for listed services (parameter -snames):
(1) run/run-registry \ (2) -e http://www.ebi.ac.uk/soaplab/services \ (3) -snames edit.seqret,display.showdb \ (4) -format udd4j \ (5) -o PublishTwoServices.java
The generated Java code is a standalone Java program that can be started and can be customized by additional arguments from the command-line. The arguments include information such as:
- what UDDI server to use
- whay user name and password to use to access the UDDI registry
- what service provider and what name for service classification should be used
And this is the relevant generated code:(1) run/run-registry \ (2) -e http://www.ebi.ac.uk/soaplab/services \ (3) -snames edit.seqret,display.showdb \ (4) -format udd4j \ (5) -o PublishTwoServices.java \ (6) -userid 'XXX' -password 'YYY' -taxonomy 'EMBOSS' -provider 'EBI'
// // parameter name default value // -------------- ------------- knownParams.put (USERID, "XXX"); knownParams.put (PASSWORD, "YYY"); knownParams.put (PUBLISHURL, "http://localhost:8080/publish"); knownParams.put (INQUIRYURL, "http://localhost:8080/inquiry"); knownParams.put (PROVIDER, "EBI"); knownParams.put (TAXONOMY, "EMBOSS");
The Taverna project aims to provide a language and software tools to facilitate easy use of workflow and distributed compute technology within the eScience community.
Its workbench is a very rich graphical user interface that is able to build and execute workflows of the components of many kinds - one of them are Soaplab services. Here is an example of a workflow similar to the EMBOSS Tutorial shown above:
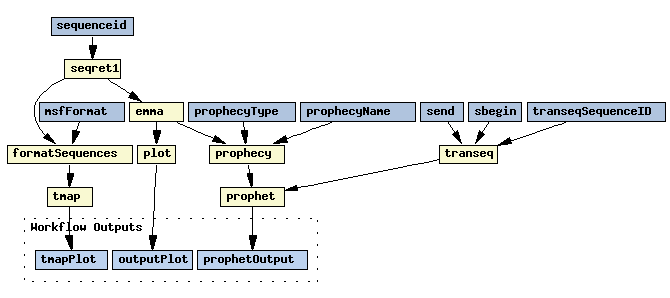
And this workflow shows that Soaplab services (backtransseq and restrict in this case) can be combined in Taverna with other kind of Web Services (BioMoby services in this case):
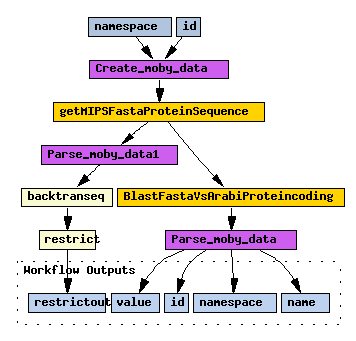
Taverna has its own very detailed user guide. Here just a quick start how to load your own Soaplab services there:
Right click on Available Processors in Available services window, and select Add new Soaplab scavenger.... Type an endpoint of your Factory (List) Service (without a service name; typically it will be something like http://localhost:8080/axis/services/) and all your services should appear.
 |
How to write your own client in Java |
Last modified: Sun Oct 7 15:08:54 2007