Soaplab provides Web Service. There are up to three types of them:
- Analysis Factory Service
- A Web Service that produces a list of available analysis, and
provides pointers to them. It does not need to be used,
the users can access individual services directly, or to use
some other (third-party) service-registry type service.
- Analysis Service
- A Web Service representing a remote analysis, a remote application.
Soaplab can deploy many of these services (there are over one
hundred of them running at EBI). Disregarding how many of them
are deployed, and what input data they expect, they all have
the same interface, they all are controlled by the same methods.
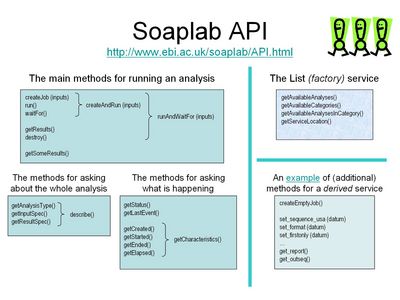
The individual services have their input data and their results named, and there are methods how to find what names are used. Once a user knows the input data names she can send her data to the analysis as the weakly-typed name-value pairs, and obtain results again as name-value pairs.
An example of using such API would be (using a pseudo-code):
Hashtable inputs = new Hashtable(); inputs.put ("sequence_direct_data", "atatatatataggcgc"); inputs.put ("osformat", "embl"); // output sequence format runAndWaitFor (inputs);Note that you need to know in advance that this service (in this example) recognizes names sequence_direct_data and osformat. - Derived Analysis Service
- A Web service representing the same remote analysis as the one above,
but with strongly-typed methods for sending input data and
receiving data results.
The same example as above but now with an API for a derived service:
Job job = createEmptyJob(); job.set_sequence_direct_data ("atatatatataggcgc"); job.set_osformat ("embl"); // output sequence format run (job); waitFor (job);Again, a site can provide many of such services. They can coexist with the non-derived analysis services (because their service names differ from the normal ones by a suffix .derived).The Soaplab allows to create implementation Java classes and the corresponding deployment descriptors for all derived analysis services automatically (Soaplab generates and compiles them during the deployment).
The same kinds of services apply to Gowlab sub-project. The only difference is on the server side where the Gowlab services do not use internally any CORBA - therefore, for Gowlab services ignore the CORBA details below.
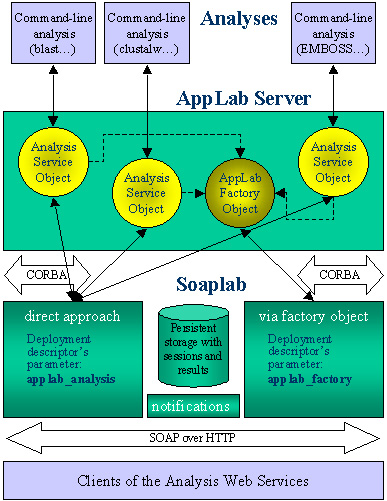
Soaplab does not access individual analysis programs directly but it uses a general-purpose package AppLab that hides all details about finding, starting, controlling, and using applications programs. The AppLab is a CORBA-based implementation of the "Biomolecular Sequence Analysis" engine specification, standardized by the Object Managememnt Group (see detail in the AppLab pages).
The advantages of AppLab (and actually of the whole specification) lie in a standard way how to describe analyses and their input and output data by an XML-based metadata description. The Soaplab Web Service interface allows accessing the metadata.
The Soaplab users do not see any CORBA communication - for them it is just an implementation details.
The Soaplab service providers can choose what AppLab objects to contact. There are two options:
The both variants and the entire architecture overview is seen in the picture:
- Use, for all services, the AppLab's AppLabFactory object:
- This alternative is easier for deploying because all deployment descriptors of all Analysis Web Services have the same value of the parameter applab_factory. It contains a location of an IOR of the AppLab's Factory object (or the IOR directly).
- Use individual AppLab's object representing individual analyses directly:
- With this alternative, use parameter applab_analysis with value different for each Analysis Web Service (containing again a location of an IOR or an IOR itself).
The short description above may be confusing unless you already have deployed Soaplab services. The detailed guides are available:
 |
How to deploy Soaplab services |
 |
How to use Soaplab services |
Additional pictures can help to understand the architecture:
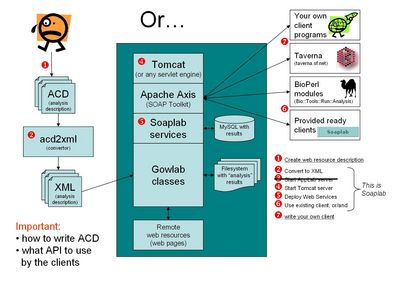
An overall view how to use Soaplab

Soaplab API
Last modified: Wed Jul 27 11:07:21 2005